In December 2016, I apparently had lots of time to kill. Two years into my PhD I had acquired enough programming and Linux skills that I felt comfortable enough to start experimenting with these credit card-size computers. So I borrowed a Raspberry Pi from a friend and started looking for possible interesting projects. I quickly became fascinated by the idea of launching and maintaining my own Twitter robot, because what child does not want to have its own robot?
But what would this robot tweet about? I have no clue on how I ended up with this idea, but I guess I was pretty obsessed with my work that I made one that would be useful for my PhD. Since I was working a lot with Lactobacillus genomes, I thought it would help me if I had something to keep track off new genomes that appeared on the NCBI Assembly database. And that is how @Lactobot was born!
Hello Twitter
— Lacto bot (@Lactobot) December 17, 2016
Due to my basic knowledge of visualising data with Python (much more of an R expert here), the first tweets of @Lactobot were extremely ugly. But it worked pretty smooth nevertheless! As soon as a new Lactobacillus genome appeared on NCBI, @Lactobot would tweet about it:
There are currently 1007 Lactobacillus assemblies available. That's 3 more than yesterday. #lactobot pic.twitter.com/JSne9yfBL9
— Lacto bot (@Lactobot) December 22, 2016
Fast forward to 2020. @Lactobot has been working for 4 years (almost) non-stop, has improved it’s data visualisation and has moved 3 times, including one International move:
Dear followers,
— Lacto bot (@Lactobot) July 26, 2019
My boss says I work to hard so I am obliged to take a holiday now.
I will take this opportunity to catch some sleep and explore other physical locations (🇧🇪 -> 🇩🇪).
I'll probably be back in a month or so, when I find a suitable internet connection.
Stay cool!
It saw the number of Lactobacillus genomes rise from 1004 to 3220:
There are currently 3220 Lactobacillus assemblies available. That's 1 more than yesterday. #lactobot pic.twitter.com/eKNYLZgxnr
— Lacto bot (@Lactobot) August 26, 2020
But then in August 2020, it started to act weirdly with a sudden drop in the total number of genomes.
There are currently 2717 Lactobacillus assemblies available. That's 6 more than yesterday. #lactobot pic.twitter.com/EpiiGo3AXy
— Lacto bot (@Lactobot) August 30, 2020
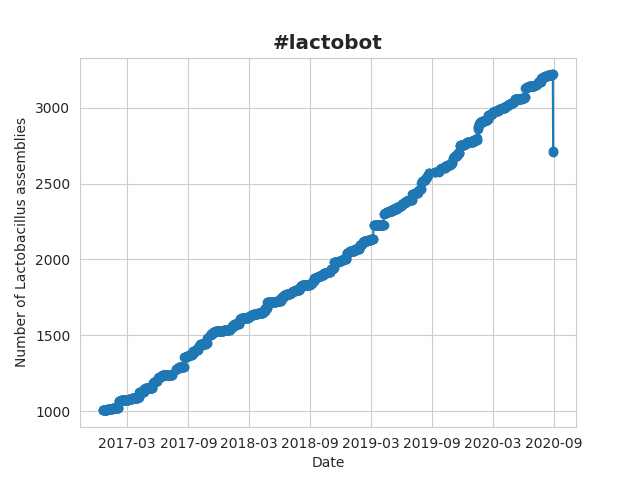
I have seen some of these drops before, but never as pronounced as this one. Usually these were small bugs and I needed to push some small updates, to get it back on track. However this time, it was clear that the problem was much bigger, and it might have been (partially) my own fault.
It all had to to with this paper we published in April 2020. In that manuscript we propose an official reclassification of the Lactobacillus genus which splits the genus into 25 smaller genera. The consequences are that currently only around 37 species have kept the original Lactobacillus name. The other (more or less) 200 species have received a complete novel genus name (Find out what name with this tool I designed in collaboration with my colleagues). And @Lactobot‘s basic code base in not good enough to properly pick up all those new names. So I basically had three options:
- Update @Lactobot to stay in line with the new taxonomy and keep track of 25 different genera.
- Reduce @Lactobot to only printing genomes related to the novel Lactobacillus genus.
- Pull the plug
Since my move to the EMBL, I have altered my research focus, and am not so much working with lactobacilli any more. Therefore, I decided it might not be worth my time any more to update this basic Twitter robot and instead move on to other projects. So my apologies to all 54 followers, but I chose for option number 3: Pull the plug and end the 4-year journey of @Lactobot.
Goodbye my dear friend

PS: If anyone else is interested in keeping this project alive, you can always contact me or check out the code on Github.
One thought on “I might have helped my own Twitter robot (@Lactobot) to its end”
R.I.P. Lactobot, you were dearly loved by us lactofans!