I am a huge fan of the statistical programming language R. The initial reason why I fell in love with R is mainly because of it’s awesome data visualisation library ggplot2. The versatility of this package makes it probably the most powerful tool in my data science/bio-informatics toolbox. Slowly, more and more packages related to ggplot2, came on my radar and before I knew it, I was addicted to the Tidyverse. (For those unfamiliar with the Tidyverse; it’s a set of R-packages that share the same philosophy and grammar. It’s great for data science). I did not have a lot of reasons to leave this great universe. It totally made me forget about Python.
As my career progressed, I learned more and more about bio-informatics, which also resulted in increasing sizes of data that had to be crunched. I must say that R did a pretty decent job in handling all of this (I even used it to decode computer files that were stored on DNA). But since I joined the Bork lab, I find myself running into its limits much faster than before. So I feel that it is time to sharpen some other knives in my toolbox. Dusting off my Python skills would be a logic next step, but then I stumbled onto something newer: Julia…
Julia: A programming language that walks like Python, but runs like C
Obligatory quote about Julia that I found somewhere on the internet
I don’t know what it is exactly that attracts me to this language (maybe it’s FOMO), but I am somewhat eager to test it out. So whenever I now need to solve a task that seems to be unbearable to be processed in R, I try to turn to Julia. And the first task was:
Extract relevant lines from large file using IDs from other file
So for this task, I would like to extract lines from a large file (32gb; File B) based on an ID (File A) that is stored in another file.
> FileA
ID01
ID02
ID03
...
IDXX
> FileB
ID05 information more_information even_more_information 102
ID12 information more_information even_more_information 115
ID89 information more_information even_more_information 009
...
And this is the first Julia script that I wrote to process these files.
# Read in query file
## Initiate empty vector
queries = Any[]
## Loop through file and populate vector with text
open("fileA") do query_file
for ln in eachline(query_file)
push!(queries, ln)
end
end
# Loop through database file
open("FileB") do file
## Loop through lines
for ln in eachline(file)
# Extract ID with regex
ID = match(r"^(\S)+", ln).match
# search in queries
if ID in queries
println(ln)
end
end
end
This was not a great success. I had this script running for more than 30 minutes and it produced nothing at all. For reference; a colleague wrote a Perl script that did the job in 12 minutes. Clearly something was wrong here. So I turned to the pro’s and asked for their advice on the Discourse forum.
One user immediately saw three red flags, the main one being: Avoid Global Variables. (See these performance tips). Based on these tips, I rearranged some code and drafted this Julia script:
# Extract IDs from query file
function get_ids(file)
## Initiate empty vector
queries = Set{String}()
## Loop through file and populate vector with text
for ln in eachline(file)
push!(queries, ln)
end
return queries
end
# Check ids with db
function compare_ids(queries, db)
# Loop through db_lines
for ln in eachline(db)
# Extract ID with regex
ID = match(r"^(\S)+", ln).match
# Search in queries
if ID in queries
println(ln)
end
end
end
# Run the actual functions
open("FileA") do query_file
queries = get_ids(query_file)
open("FileB") do db_file
compare_ids(queries, db_file)
end
end
It is not the most beautiful piece of code that I wrote, but boy, was I happy with its execution time!
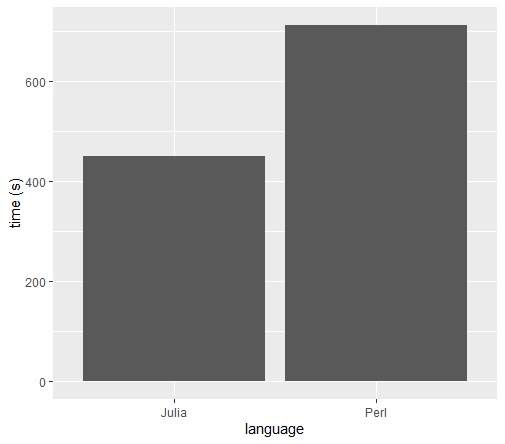
To be honest, the above “benchmark” might not be super fair since my colleague spent 5 minutes writing his Perl script, while it took me several rounds to optimise my Julia version. Nevertheless, I have successfully performed my first baby steps into Julia and have confidence it can make quite some impact in future analyses.
But to achieve that, I will definitely need more practice!
Cheers,
Sander
PS: For completeness, here is the Perl script:
open(IN, 'FileA');
while (<IN>) {
chomp;
$i_need{$_}++;
}
open (IN, 'FileB') ;
while (<IN>) {
/^(\S+)\t/;
if ($i_need{$1}) { print; }
}
2 thoughts on “Introducing myself to Julia”
If all the IDs are exactly two digits, you can solve the problem much faster (no coding at all and faster runtime) by just using this standard Unix command:
grep -f FileA FileB
If the IDs have varying length, the problem becomes a little bit more complex, because one digit-sequence can be a subset of another, but the general idea remains: if the problem is pattern matching in a big text file, just use grep. It has many options and is using the fast (sub-linear) Boyer Moore algorithm.
Thanks for passing by Ariel,
I am familiar with grep and because of the substring matching, this was not an option. In reality the ID’s were also much longer and more complex than what I described in the example. This would thus definitely lead to a faulty result. In addition, I was surprised that it also had a long runtime on my first test (grep was the first thing I tried), it did not finish on the fly.
Finally, I would like to point out, as stated in the beginning of the blog post, that this was not so much about finding the fastest solution, but more about “Introducing myself to Julia”.